Modular Platform
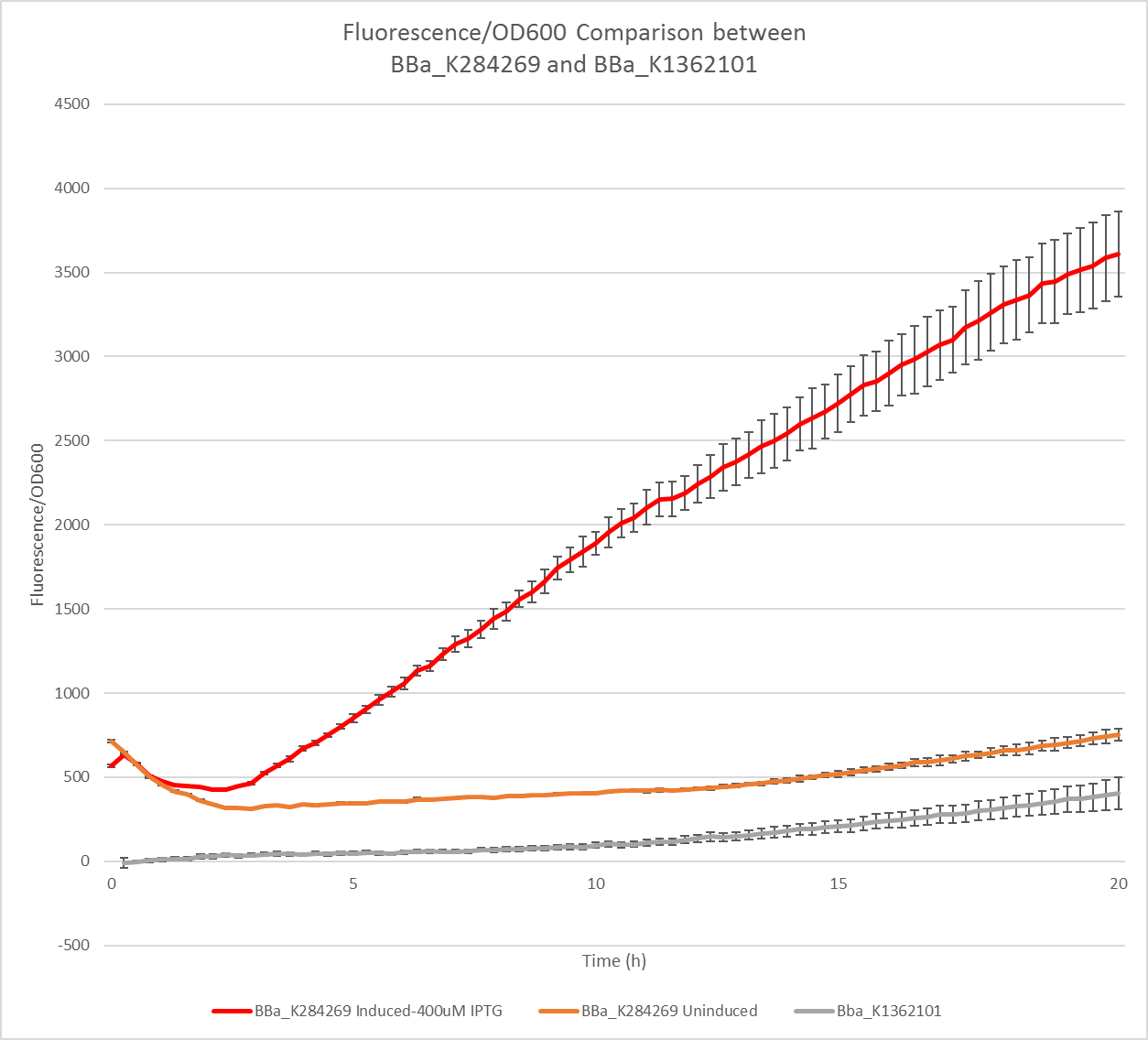
Starting from the an idea centered around spider silk and its applications, after an insightful conversation with the CSO of Bolt Threads, we devised and developed a tunable construct which brings modular protein engineering in the realm of biomaterials. First, we considered using the spytag-spycatcher system, but we realised that there would have been a much better method to reach our objective. Spliced inteins contribute to the assembly of fusion proteins through directed covalent bonding, in the same fashion as spytag and spycatcher[1]. However, spytag and spycatcher risk to influence the folding and polymerisation process of biomaterials regulated by tight structural constraints through steric hindrance due to their size. Split-inteins avoid this type of issue by splicing out and leaving a permanent covalent bond in a short time, which varies slightly according to literature[2]. In order to simulate the association and potential polymerization through split inteins, we designed a green-red split-inteins system which alternates well characterised chromogenic proteins: GFP, and RFP [3]. Additionally, we devised an Intein Passenger construct to host any peptide of interest to functionalise the biopolymer characterised by an mScarlet report. We obtained plate reader data on the fluorescence of our mScarlet, which confirmed its superiority as an efficient reporter, especially if compared with another intein reporter construct uploaded on the registry by Heidelberg’s iGEM team in 2014.
BioBrick 2.0
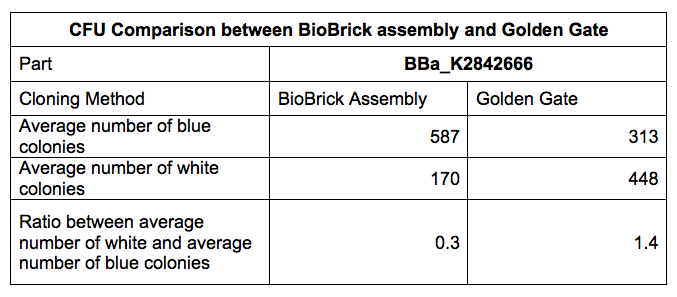
Having singled out time constraints as the most concerning issue for the majority of iGEM teams, we designed a shortcut to solve this problem and increase design flexibility. Our proposed BioBrick2.0 enables cloning through BioBrick, Golden Gate and Gibson assembly. We demonstrated this through a colony forming unit experiment comparing the average number of white to blue colonies between BioBrick and Golden Gate assembly (Table 1).
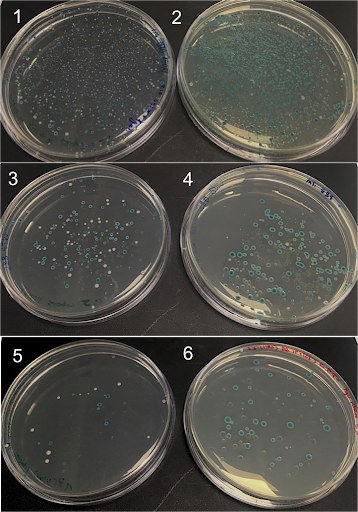
(1) Golden Gate Assembly undiluted
(2) BioBrick Assembly undiluted
(3) Golden Gate Assembly 10x dilution
(4) BioBrick Assembly 10x dilution
(5) Golden Gate Assembly 100x dilution
(6) BioBrick Assembly 100x dilution
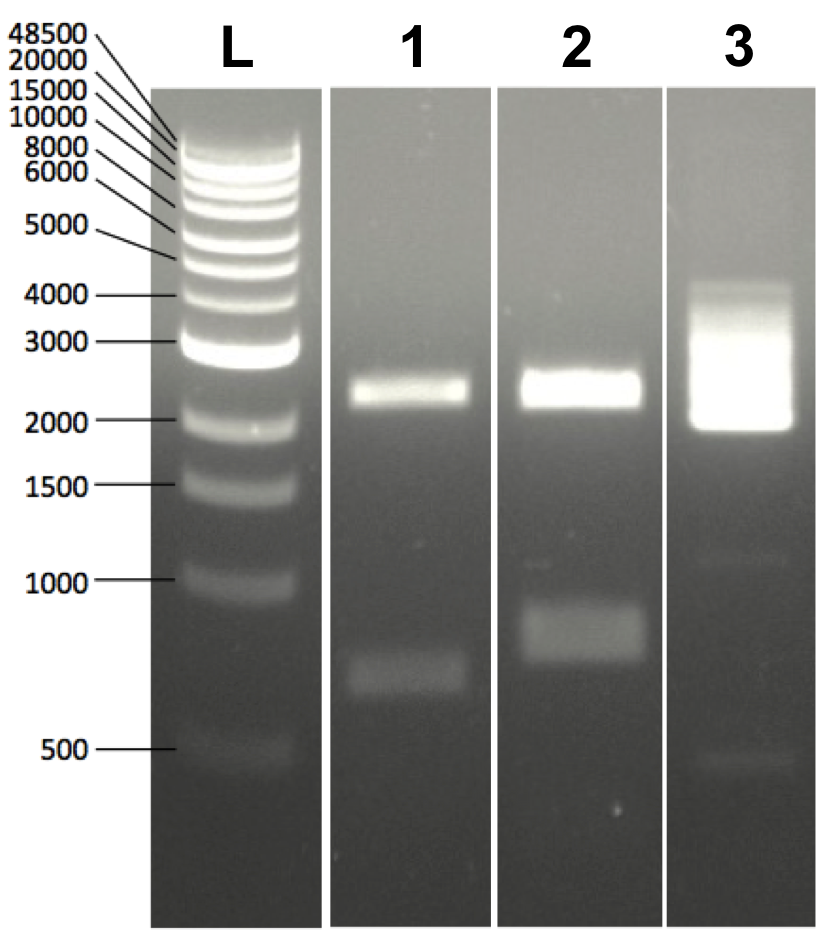
(1) BsaI digested BioBrick2.0 (Golden Gate Assembly)
(2) EcoRI+PstI digested BioBrick2.0 (BioBrick Assembly)
(3) undigested BioBrick2.0
Integrated Human Practices
The benefits of our project on society would come from direct interactions with the manufacturing industry. If spider silk polymerisation and functionalisation could be made possible but also cheaper, it would reflect on the cost of production and hence selling. This is why we decided to target Bolt Threads first which gave us a huge insight on the biomaterials industry. This impacted on the way we thought about and designed our project. Our experiments were based on spider silk alone initially but we ended up inventing a modular platform for all biomaterials in the end. This tunable platform is made of 3 main parts which can be made to plug-and-play with different biomaterials. We changed our experiments to polymerise other proteins rather than spider silk alone. Other conversations made us look into the way we’d apply a spider silk filter in a community and other functionalizing proteins we could apply to our biomaterials.
Integrated Modelling
We integrated all of our models into our work - the first kinetic model we looked at how varying IPTG concentrations and cell broth would affect gene and protein expression as we scaled up our project. The result of our first model is using 500 mM of IPTG which the model suggests as the most efficient option, and choosing TB as the medium with better carrying capacity.
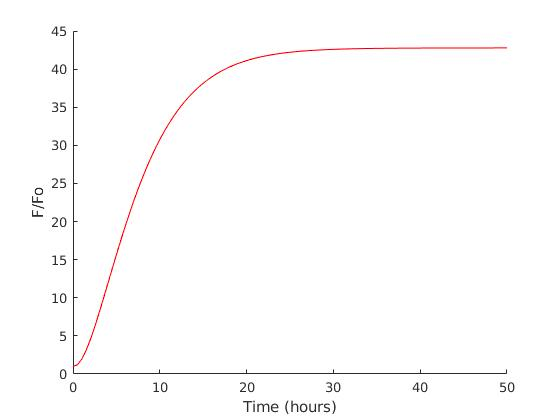
We found that the maximum protein fold-change as 42.8 ratio over control. It can be achieved by inoculating the cells just over 25 hours. This is really important to know the maximum yield of the process and even determine the efficiency of the protein extraction in the next steps.
The second model is the intein polymerisation, it gives us the estimate time required to produce the certain length of polymer before doing the actual experiment. From the results using specifically our intein, we then know 15 minutes are enough time to get about 90% of minimum polymer species 12 (dodecamer). Then molecular dynamics simulations are done to give an insight on the structure of our protein fiber to determine the desired polymer length to achieve good mechanical properties, the result is then fed back into the BioBrick design so we are able to construct the polymer without doing an actual lab experiment and hence reduce unnecessary steps, time and energy spent.




